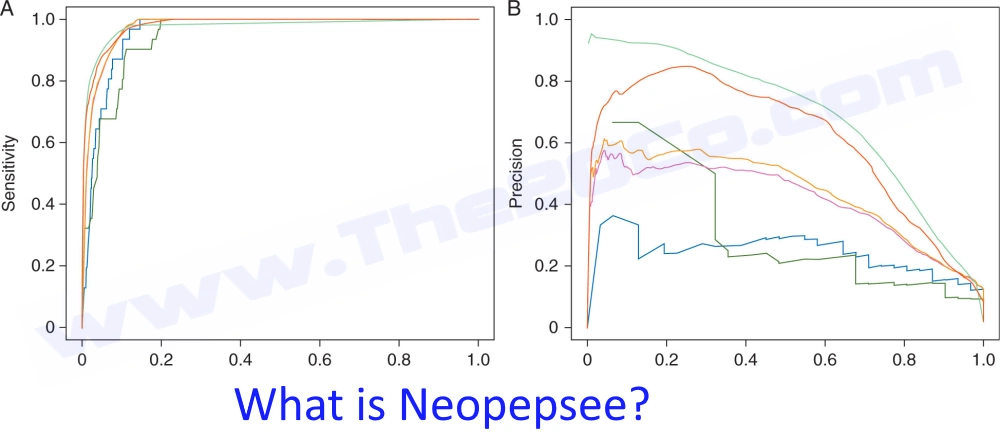
Neopepsee (Neutralizing Epitopes of Protein/Enzyme Sequence Estimator) is a newly developed algorithm for immune peptide discovery that specializes in predicting and providing annotated sequences of protein/enzyme candidate peptides and their positional values, which are crucial information for subsequent epitope mapping experiments.
What is Neopepsee?
Neopepsee is a tool for accurate genome-level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. It uses the antigenic index (AI) and the Kallman-Nelson index (KNI) to predict antigenic sites in a given protein sequence.
Neopepsee provides an API that allows you to submit your own protein sequences, which can be single or multiple, to predict their neoantigen potential.
How does Neopepsee work?
Neopepsee (Neoantigen Prediction by Sequences Extracted from Epitopes) is a bioinformatics tool designed to predict individual neoantigens in cancer genomes. Neoantigens are antigenic peptides that are derived from the genomic rearrangements of cancer cells. The most common neoantigens in cancer are somatic mutations that arise during tumor progression and can be found in multiple mutated genes. These neoantigens can have different sequences or lengths, which makes them difficult to predict with existing methods.
Neopepsee predicts unique neoantigens that are not present in the human genome reference sequence Hg19 and are predicted to be immunogenic based on their amino acid content and sequence features. Neopepsee also predicts novel alleles of known genes that have not been identified previously as being altered during tumor progression.
Detects neoantigen candidates with less false positives
The first step in the identification of a neoantigen is to find a tumor-specific protein. The most common method for this is to use a neopeptide library, which has been generated by screening peptides against a panel of cancer tissue samples.
The second step is to identify the cleavage sites between the immunogenic parts from the non-immunogenic parts. This can be done using deFuse-Trinity or other algorithms that are capable of finding immunogenic peptides.
The final step is to identify whether there are any known fusion proteins that can be used as neoantigens. If there are none, then you can try and find a new fusion protein that will make an excellent vaccine candidate.
Provides rich annotation of candidate peptides
A new database called Neopepsee provides a wealth of information on candidate peptides identified in circulating tumor cells (CTCs). The database contains over 300,000 reference sequences, including canonical peptides, non-canonical peptides, and Chinese peptides. It can be searched by cancer type, HLA allele, or immunopeptidome profile.
Neopepsee is particularly useful for those who are working with melanoma. It includes a search tool for class I and II neoantigens and also offers an option to search for candidate peptides from CTCs. This makes it possible to identify neoantigens that may be important in the development of personalized cancer vaccines or immune checkpoint therapy.
The database also contains links to other valuable resources such as dbPepNeo2.0 and the International Cancer Genome Consortium (ICGC) portal.
Predicts 7 neoantigens per patient
When it comes to cancer immunotherapy, predicting which patients will respond to treatment is crucial. Certain mutations in the tumors can indicate whether or not immune therapy will work. This can be determined by studying the mutations in the tumor and looking for those that are “neoantigens,” which means the immune system has never seen them before. The new generation of vaccines uses a multimodal approach that combines data from somatic variants, RNA expression, and the immune system’s response to predict epitopes that are useful in vaccines.
To study the expression of mutant proteins in cancer, scientists use RNA sequencing, which analyzes RNA sequences. While RNA sequencing is useful for detecting changes in expression, scientists can also combine it with DNA sequencing to identify mutations. The process for identifying neoantigens involves a lot of steps. A computer must first identify mutated DNA. Then, it must process the mutated DNA into peptides. Finally, it must identify possible binding sites for these peptides on the MHC molecule.
There are many ways to predict neoantigens. These methods are all useful, but the best ones give you a lot of information. First, they tell you the genomic location of the neoantigen. They also tell you the ClinGen ID number and the Human Genome Variation Society variant name for that gene. The prediction pipeline also gives you an indication of where within the T cell receptor (TCR) binding site the neoantigen lies.
Result of Neopepsee
The neoepitope prediction tool Neopepsee is an open-source tool that can be used to predict neoantigens from patient-derived immune repertoire data. The tool was developed by the Institute for Computational Molecular Biology (ICMB) at the Helmholtz Zentrum München (HMGU). In this study, we use a machine learning approach to build a classifier of neoantigens based on immunogenicity features. Nine of the 14 immunogenicity features that are informative and inter-independent were used to construct the machine-learning classifiers.
Neopepsee provides a rich annotation of candidate peptides with 87 immunogenicity-related values, including IC50, expression levels of neopeptides and immune regulatory genes (e.g. PD1, PD-L1), matched epitope sequences, and a three-level (high, medium, and low) call for neoantigen probability. Compared with the conventional methods, the performance was improved in sensitivity and especially two- to threefold in the specificity. Tests with validated datasets and independently proven neoantigens confirmed the improved performance in melanoma and chronic lymphocytic leukemia.
Features of Neopepsee
The following features have been implemented:
- Neopepsee uses a combination of sequence and amino acid sequence features to predict antigens with high accuracy.
- The prediction results are easily interpretable and provide detailed information about each predicted antigen, including its location, length, composition, and degree of similarity with other known antigens in the database.
- Neopepsee provides a web interface that allows users to upload their own protein sequences as well as download the predicted antigens in a FASTA format for further analysis using other tools such as eHive or JBrowse.
- Neopepsee is a machine learning method that predicts which protein sequences will become neoantigens through sequence analysis and amino acid immunogenicity analysis.
Conclusion
Neopepsee is an advanced, next-generation candidate neoantigen detection platform. The primary advantage of the technology comes from its improved detection method.
NeoPepsee allows researchers to efficiently determine potential neoantigen identities, which can be useful in developing personalized immunotherapy and vaccine treatments.